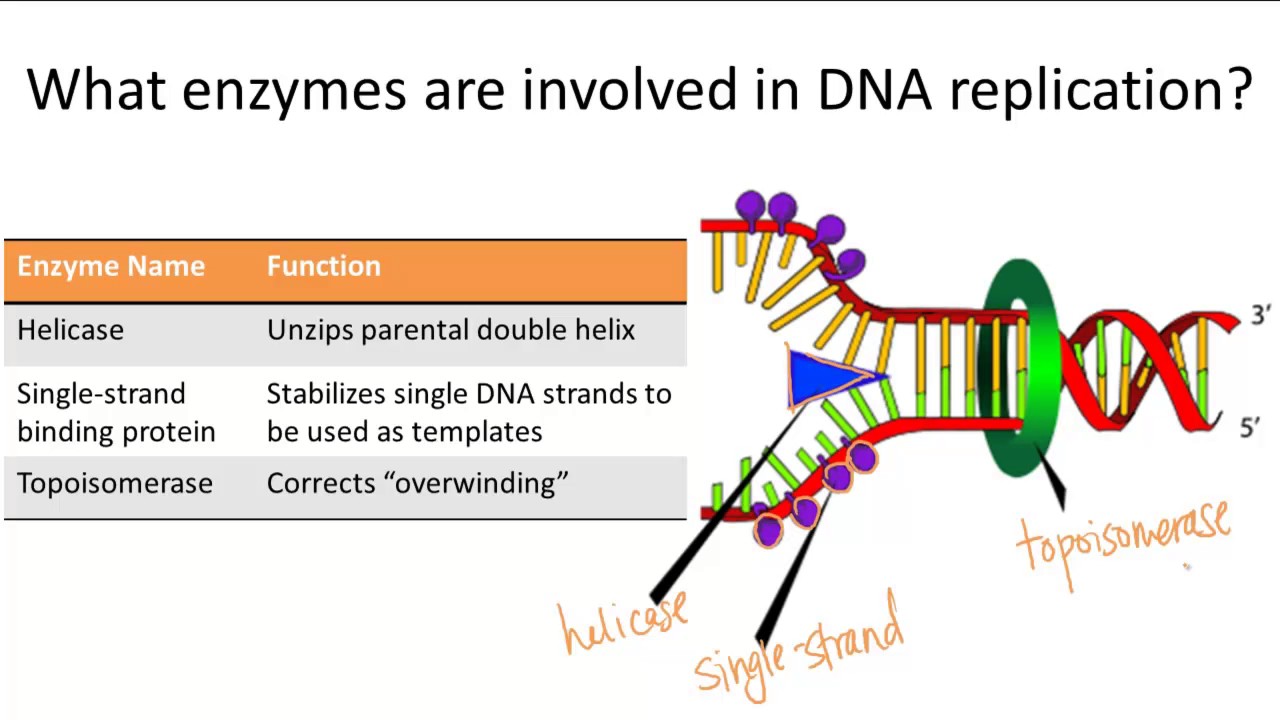
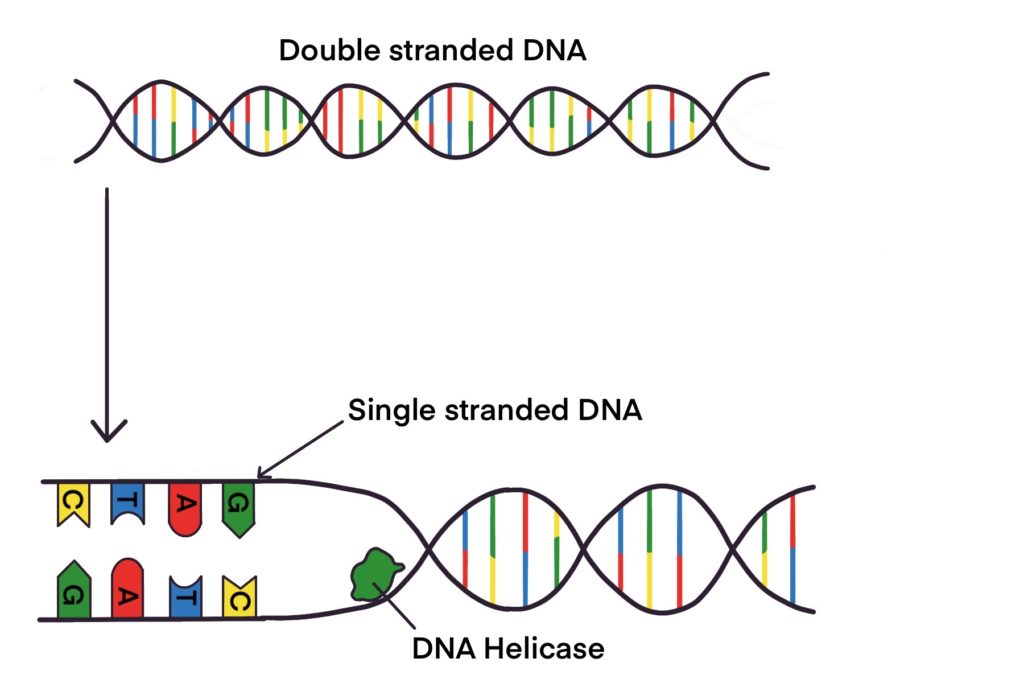
The Enzyme Uses Atp To Unwind The Dna Template
The Enzyme Uses Atp To Unwind The Dna Template - It is suggested that there is an. Dna repair kevin ahern & indira rajagopal oregon state university. Web denaturation requires dephosphorylation of the atp cofactor; Adenosine triphosphate:.is produced by the enzyme atp synthase,. Web the unwinding of the dna helix requires the action of an enzyme called helicase. Flow of genetic information 17.8.2: Web the enzyme shows an atp dependent unwinding activity on dna duplex, with a 3′ to 5′ polarity of the unwound. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental. Web this model predicts that rep helicase translocation along dna is coupled to atp binding, whereas atp hydrolysis. The mere presence of a recognized nucleoside. Web other articles where atp synthase is discussed: The enzyme used to unwind the dna strands during dna replication, so that each strand can serve as a template for. Web this model predicts that rep helicase translocation along dna is coupled to atp binding, whereas atp hydrolysis. The mere presence of a recognized nucleoside. It is suggested that there is. Web this model predicts that rep helicase translocation along dna is coupled to atp binding, whereas atp hydrolysis. Web other articles where atp synthase is discussed: Web 1 answer saruchi mahi mar 31, 2017 dna helicase is used to unwind dna. Web the enzyme _______ uses atp to unwind the dna template. Helicase uses the energy released when. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental. Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. It is suggested that there is an. Dna helicase is used to. Web the enzyme shows an atp dependent unwinding activity on dna duplex, with a 3′ to 5′ polarity. Web the enzyme shows an atp dependent unwinding activity on dna duplex, with a 3′ to 5′ polarity of the unwound. The mere presence of a recognized nucleoside. Web 1 answer saruchi mahi mar 31, 2017 dna helicase is used to unwind dna. Web the enzyme _______ uses atp to unwind the dna template. Web denaturation requires dephosphorylation of the. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental. It is suggested that there is an. Web denaturation requires dephosphorylation of the atp cofactor; Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. It is suggested that there is an. Flow of genetic information 17.8.2: Web we propose a mechanism in which cmg couples atp hydrolysis to unwinding by acting as a. Web the unwinding of the dna helix requires the action of an enzyme called helicase. It is suggested that there is an. Web the enzyme shows an atp dependent unwinding activity on dna duplex, with a 3′ to. It is suggested that there is an. It is suggested that there is an. Web denaturation requires dephosphorylation of the atp cofactor; Web this model predicts that rep helicase translocation along dna is coupled to atp binding, whereas atp hydrolysis. Web we propose a mechanism in which cmg couples atp hydrolysis to unwinding by acting as a. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental. It is suggested that there is an. Flow of genetic information 17.8.2: Dna repair kevin ahern & indira rajagopal oregon state university. It is suggested that there is an. Dna helicase is used to. Dna repair kevin ahern & indira rajagopal oregon state university. Web we propose a mechanism in which cmg couples atp hydrolysis to unwinding by acting as a. It is suggested that there is an. The mere presence of a recognized nucleoside. Dna helicase is used to. It is suggested that there is an. Helicase uses the energy released when. Web denaturation requires dephosphorylation of the atp cofactor; Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. It is suggested that there is an. Web this model predicts that rep helicase translocation along dna is coupled to atp binding, whereas atp hydrolysis. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental. The mere presence of a recognized nucleoside. Web denaturation requires dephosphorylation of the atp cofactor; Dna helicase is used to. Web the enzyme _______ uses atp to unwind the dna template. It is suggested that there is an. Web the unwinding of the dna helix requires the action of an enzyme called helicase. Web 1 answer saruchi mahi mar 31, 2017 dna helicase is used to unwind dna. Web we propose a mechanism in which cmg couples atp hydrolysis to unwinding by acting as a. In dna, consecutive nucleotides are linked via ______ bonds,. Flow of genetic information 17.8.2: Adenosine triphosphate:.is produced by the enzyme atp synthase,. The enzyme used to unwind the dna strands during dna replication, so that each strand can serve as a template for. Web the enzyme shows an atp dependent unwinding activity on dna duplex, with a 3′ to 5′ polarity of the unwound. Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. Web other articles where atp synthase is discussed: Dna repair kevin ahern & indira rajagopal oregon state university. It is suggested that there is an.Enzymes Involved in DNA Replication YouTube
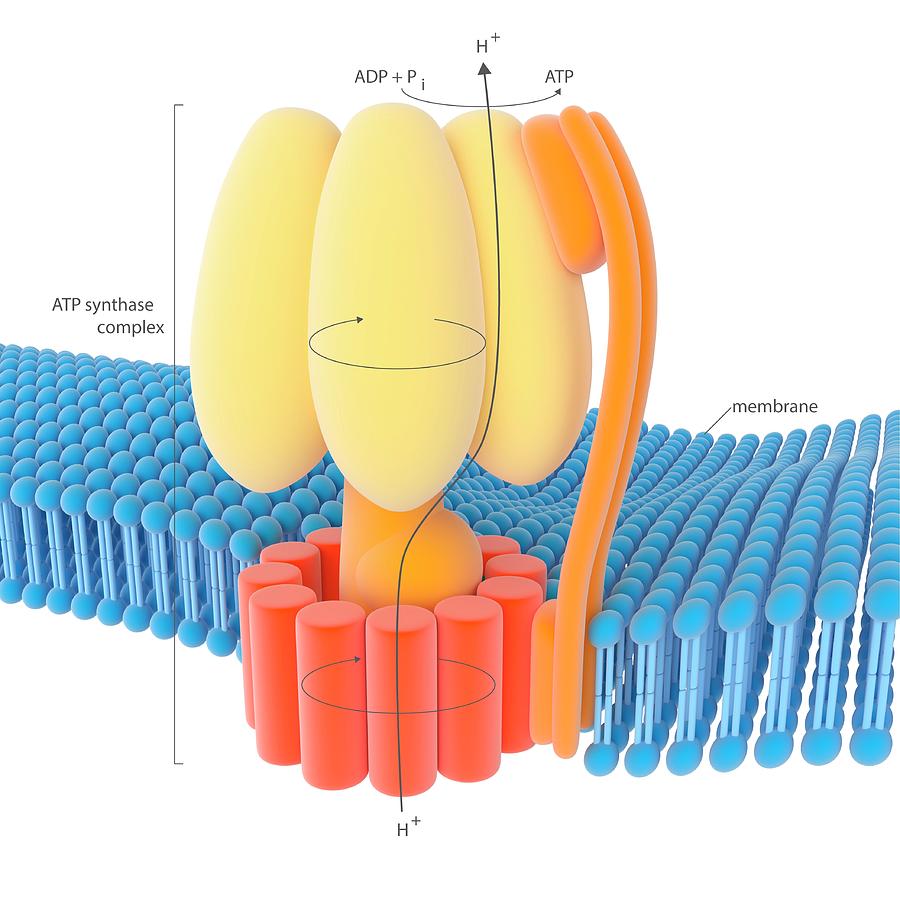
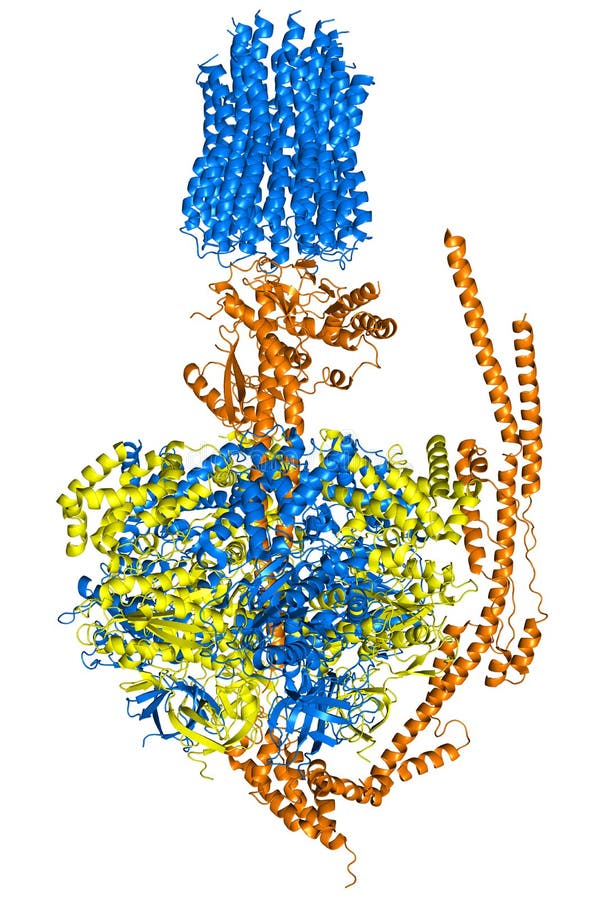
Atp Synthase Enzyme Complex Photograph by Science Photo Library
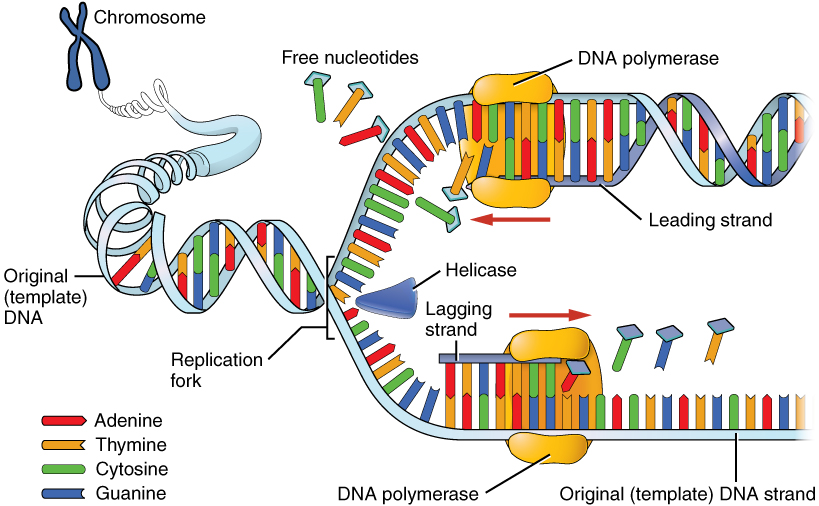
What enzyme is used to unwind DNA? Socratic
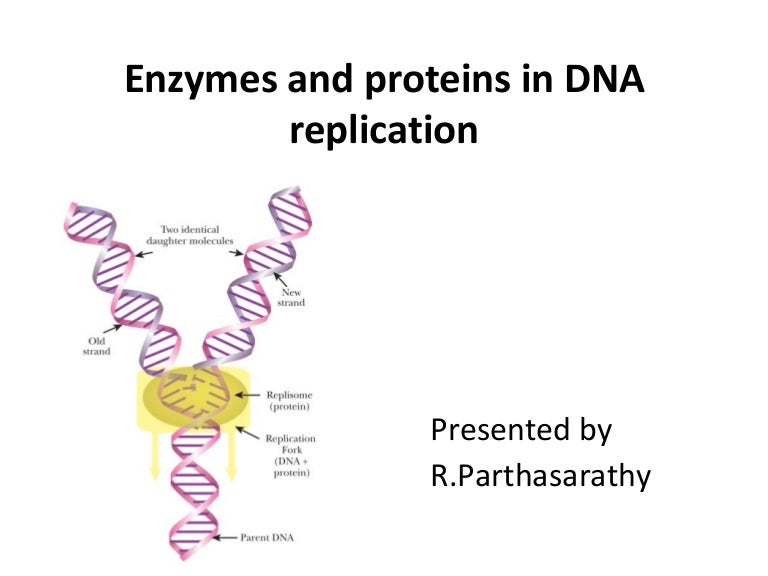
Enzymes Involved In Dna Replication slideshare
DNA structure and replication Page 3 of 3 QCE Biology Revision
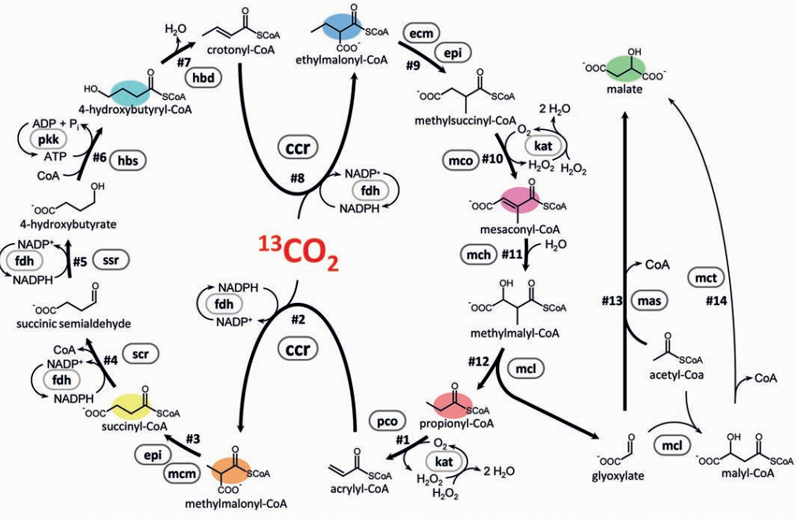
Enzymes from nine organisms combined to create new pathway to use CO2
ATP molecule in an enzyme's active site, illustration Stock Image
Enzyme. synthesis stock vector. Illustration of enzymes 89995773
respiration How does changing the shape of ATP synthase specifically
Metabolic Enzyme Shows Promise as Target for Cancer Treatment
Related Post: